home — research — team — resources — lab news
Our research
We focus on single-molecule experiments in living cells using fluorescent proteins and other fluorescent molecules.
Please contact zach.hensel@itqb.unl.pt if you are interested in working on a project in the lab or collaborating. Two of the possible themes for a research project are imaging transcription elongation in real time with single-molecule resolution and investigating how mycobacterial cell division responds to environmental stress both with fluorescence microscopy and computational methods.
We also work on improved methods for recombinant gene expression and have distributed some plasmids that are very useful for low-noise gene expression that can be found at AddGene.
Publications from lab members
Google Scholar - ORCID: 0000-0002-4348-6229
Lab members in bold
† Equal contribution
* Corresponding author
# Preprint not published elsewhere
Research manuscripts (including preprints not published elsewhere)
Click titles to expand abstracts and see data/analysis from our lab for papers since 2022!
Z Hensel*, F Débarre. An updated dataset of early SARS-CoV-2 diversity supports a wildlife market origin. bioRxiv 647275, 2025. https://doi.org/10.1101/2025.04.05.647275
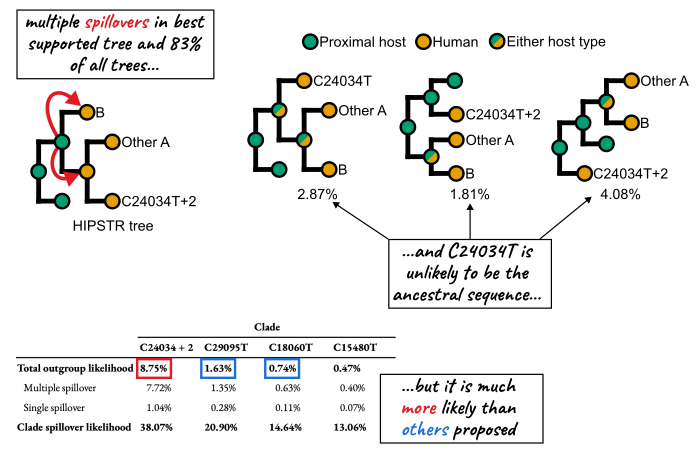
Abstract: The origin of SARS-CoV-2 has been intensely scrutinized, and epidemiological and genomic evidence has consistently pointed to Wuhan’s Huanan Seafood Wholesale Market as the epicenter of the COVID-19 pandemic. Early cases were associated with this market, and environmental sequencing placed the common ancestor of SARS-CoV-2 genomic diversity within the market. Phylogenetic analysis also suggested separate introductions of lineages A and B into the human population, a finding that can be tested with additional data. Here, we curated an expanded sequence dataset of early SARS-CoV-2 viral genomes, including newly available sequences from mid-January 2020. In this dataset, we found no additional support for previously proposed alternative progenitor sequences, or for any evolutionary intermediates between lineages A and B in the human population. Instead, we identified SARS-CoV-2 lineages that may have spread from the market, and additional samples of a sublineage of lineage A with three mutations, including one found in closely related bat coronaviruses. Although our analysis of early pandemic genomes suggests that this mutation is unlikely to characterize the immediate SARS-CoV-2 ancestor, it is more plausible than two previously proposed ancestral genomes. These findings reinforce the proposed emergence of SARS-CoV-2 from the wildlife trade at the Huanan market, demonstrating how new data continues to both solidify and clarify our understanding of how the pandemic began.
†JR Ferreira, †R Xu, Z Hensel*. Mycobacterium tuberculosis FtsB and PerM interact via a C-terminal helix in FtsB to modulate cell division. Journal of Bacteriology 207, 2025. https://doi.org/10.1128/jb.00444-24
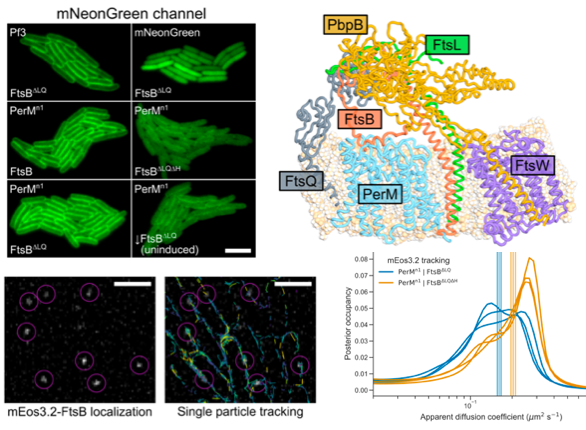
Abstract: Latent infection by Mycobacterium tuberculosis (Mtb) impedes effective tuberculosis therapy and eradication. The protein PerM is essential for chronic Mtb infections in mice and acts via the divisome protein FtsB to modulate cell division. Using transgenic co-expression in Escherichia coli, we studied the Mtb PerM-FtsB interaction in isolation from other Mtb proteins, engineering PerM to enhance expression in the E. coli membrane. We confirmed the reported instability of Mtb FtsB, and we linked FtsB instability to a segment of FtsB predicted to bind cell-division proteins FtsL and FtsQ. Though narrowly conserved, the PerM-FtsB interaction emerges as a potential target for therapy targeting persistent infections by disrupting regulation of cell division. Using fluorescence microscopy, we found that stability of both FtsB and PerM hinges on their interaction via a C-terminal helix in FtsB. Molecular dynamics results supported the observation that FtsB stabilized PerM, and suggested that interactions at the PerM-FtsB interface differ from our initial structure prediction in a way that is consistent with PerM sequence conservation. Integrating protein structure prediction, molecular dynamics and single-molecule microscopy, our approach is primed to screen potential inhibitors of the PerM-FtsB interaction and can be straightforwardly adapted to explore other putative interactions.
JE Pekar*, N Moshiri, P Lemey, A Crits-Christoph, F Débarre, SA Golstein, Z Hensel, A Rambaut, M Worobey, EC Holmes, JO Wertheim*. Recently reported SARS-CoV-2 genomes suggested to be intermediate between the two early main lineages are instead likely derived. Virus Evolution 11, 2025. https://doi.org/10.1093/ve/veaf008
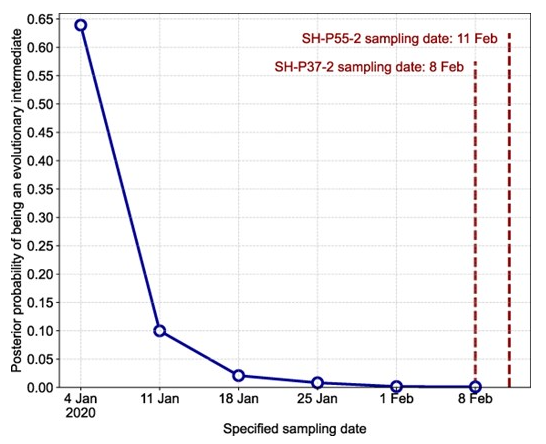
Abstract: Understanding the genomic diversity of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) at the outset of the coronavirus disease 2019 pandemic can provide insight into the circumstances leading to its emergence. Early SARS-CoV-2 genomic diversity has been classified into two distinct viral lineages, denoted “A” and “B,” which we hypothesized were separately introduced into humans. Recently published data contain two genomes with a haplotype suggested to be an evolutionary intermediate to these two lineages, known as “T/T.” We used a phylodynamic approach to analyze SARS-CoV-2 genomes from early 2020 to determine whether these two T/T genomes represent an evolutionarily intermediate haplotype between lineages A and B, or if they are a later descendent of either of these two lineages. We find that these two recently published T/T genomes do not represent an evolutionarily intermediate haplotype and were, instead, derived from either lineage A or lineage B. However, we cannot conclusively determine from which lineage they were derived. After including additional data from the start of the pandemic, including these two T/T genomes, we again find a discrepancy in the molecular clock when inferring the ancestral haplotype of SARS-CoV-2, corroborating existing evidence for the separate introductions of SARS-CoV-2 lineages A and B into the human population in late 2019.
Z Hensel*. Secondary structure of the SARS-CoV-2 genome is predictive of nucleotide substitution frequency. eLife 13:RP98102, 2025. https://doi.org/10.7554/eLife.98102.3
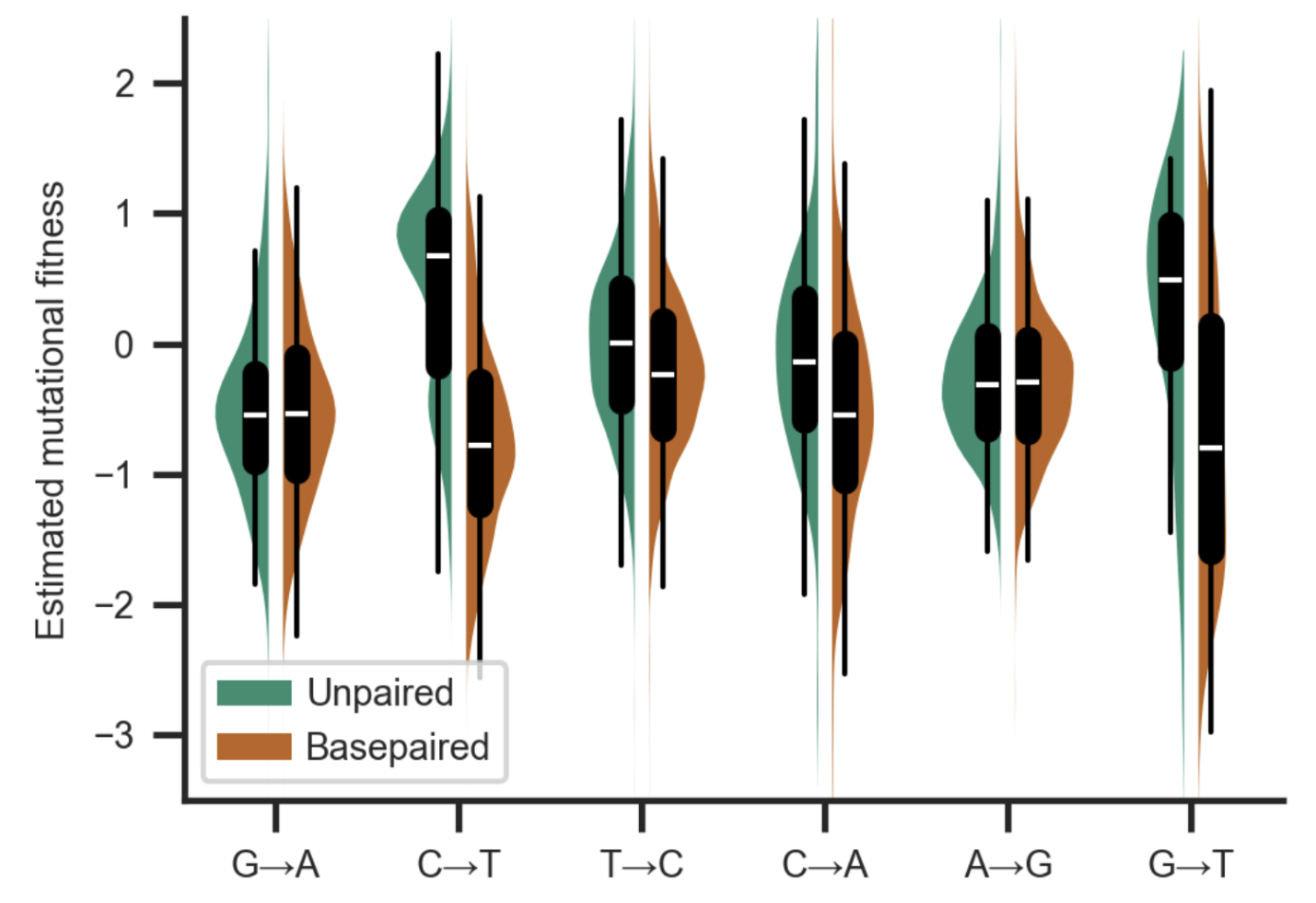
Abstract: Accurate estimation of the effects of mutations on SARS-CoV-2 viral fitness can inform public-health responses such as vaccine development and predicting the impact of a new variant; it can also illuminate biological mechanisms including those underlying the emergence of variants of concern. Recently, Lan et al reported a high-quality model of SARS-CoV-2 secondary structure and its underlying dimethyl sulfate (DMS) reactivity data. I investigated whether secondary structure can explain some variability in the frequency of observing different nucleotide substitutions across millions of patient sequences in the SARS-CoV-2 phylogenetic tree. Nucleotide basepairing was compared to the estimated “mutational fitness” of substitutions, a measurement of the difference between a substitution’s observed and expected frequency that is correlated with other estimates of viral fitness. This comparison revealed that secondary structure is often predictive of substitution frequency, with significant decreases in substitution frequencies at basepaired positions. Focusing on the mutational fitness of C→T, the most common type of substitution, I describe C→T substitutions at basepaired positions that characterize major SARS-CoV-2 variants; such mutations may have a greater impact on fitness than appreciated when considering substitution frequency alone.
# CC Santos, N Schweizer, F Cairrão, JR, Nerea Osinalde, M Yang, CJ Gaspar, VI Rasheva, Z Hensel, C Adrain, TN Cordeiro, F Voigt, PA Gameiro, U Mayor, PM Domingos*. Fbxo42 promotes the degradation of Ataxin-2 granules to trigger terminal Xbp1 signaling. bioRxiv, 2024. https://doi.org/10.1101/2024.12.22.629979
Abstract: The Unfolded Protein Response (UPR) is composed by homeostatic signaling pathways that are activated by the accumulation of misfolded proteins in the Endoplasmic Reticulum (ER), a condition known as ER stress. Prolonged ER stress and activation of the UPR causes cell death, by mechanisms that remain poorly understood. Here, we report that regulation of Ataxin-2 by Fbxo42 is a crucial step during UPR-induced cell death. From a genetic screen in Drosophila, we identified loss of function mutations in Fbxo42 that suppress cell death and retinal degeneration induced by the overexpression of Xbp1spliced, an important mediator of the UPR. We identified the RNA binding protein Ataxin-2 as a substrate of Fbxo42, which, as part of a Skp-A/Cullin-1 complex, promotes the ubiquitylation and degradation of Ataxin-2. Upon ER-stress, the mRNA of Xbp1 is not immediately translated but instead it is sequestered and stabilized in Ataxin-2 granules, until the Fbxo42 recruitment to these granules promotes the degradation of Ataxin-2, allowing the translation of Xbp1 mRNA at later stages of UPR activation. Our results identify Fbxo42-mediated degradation of Ataxin-2 as a key mechanism regulating the levels of Xbp1 mRNA and protein, to trigger cell death during the terminal stages of UPR activation.
A Crits-Christoph, JI Levy, JE Pekar, SA Goldstein, R Singh, Z Hensel, K Gangavarapu, MB Rogers, N Moshiri, RF Garry, EC Holmes, MPG Koopmans, P Lemey, S Popescu, A Rambaut, DL Robertson, MA Suchard, JO Wertheim, AL Rasmussen, KG Andersen*, M Worobey*, F Débarre*. Genetic tracing of market wildlife and viruses at the epicenter of the COVID-19 pandemic. Cell, 2024. https://doi.org/10.1016/j.cell.2024.08.010
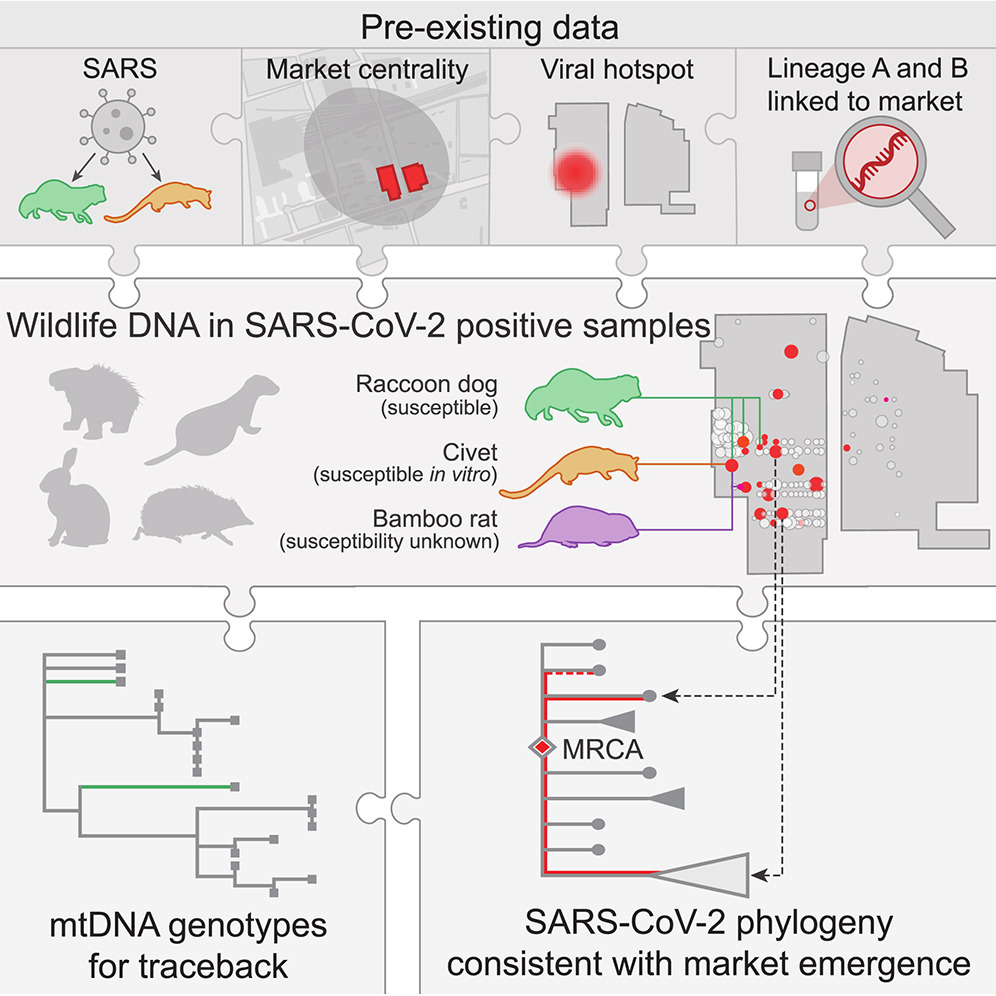
Abstract: Zoonotic spillovers of viruses have occurred through the animal trade worldwide. The start of the COVID-19 pandemic was traced epidemiologically to the Huanan Seafood Wholesale Market. Here, we analyze environmental qPCR and sequencing data collected in the Huanan market in early 2020. We demonstrate that market-linked severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genetic diversity is consistent with market emergence and find increased SARS-CoV-2 positivity near and within a wildlife stall. We identify wildlife DNA in all SARS-CoV-2-positive samples from this stall, including species such as civets, bamboo rats, and raccoon dogs, previously identified as possible intermediate hosts. We also detect animal viruses that infect raccoon dogs, civets, and bamboo rats. Combining metagenomic and phylogenetic approaches, we recover genotypes of market animals and compare them with those from farms and other markets. This analysis provides the genetic basis for a shortlist of potential intermediate hosts of SARS-CoV-2 to prioritize for serological and viral sampling.
S Schäper, AD Brito, BM Saraiva, GR Squyres, MJ Holmes, EC Garner, Z Hensel, R Henriques, MG Pinho*. Cell constriction requires processive septal peptidoglycan synthase movement independent of FtsZ treadmilling in Staphylococcus aureus. Nature Microbiology, 2024. https://doi.org/10.1038/s41564-024-01629-6
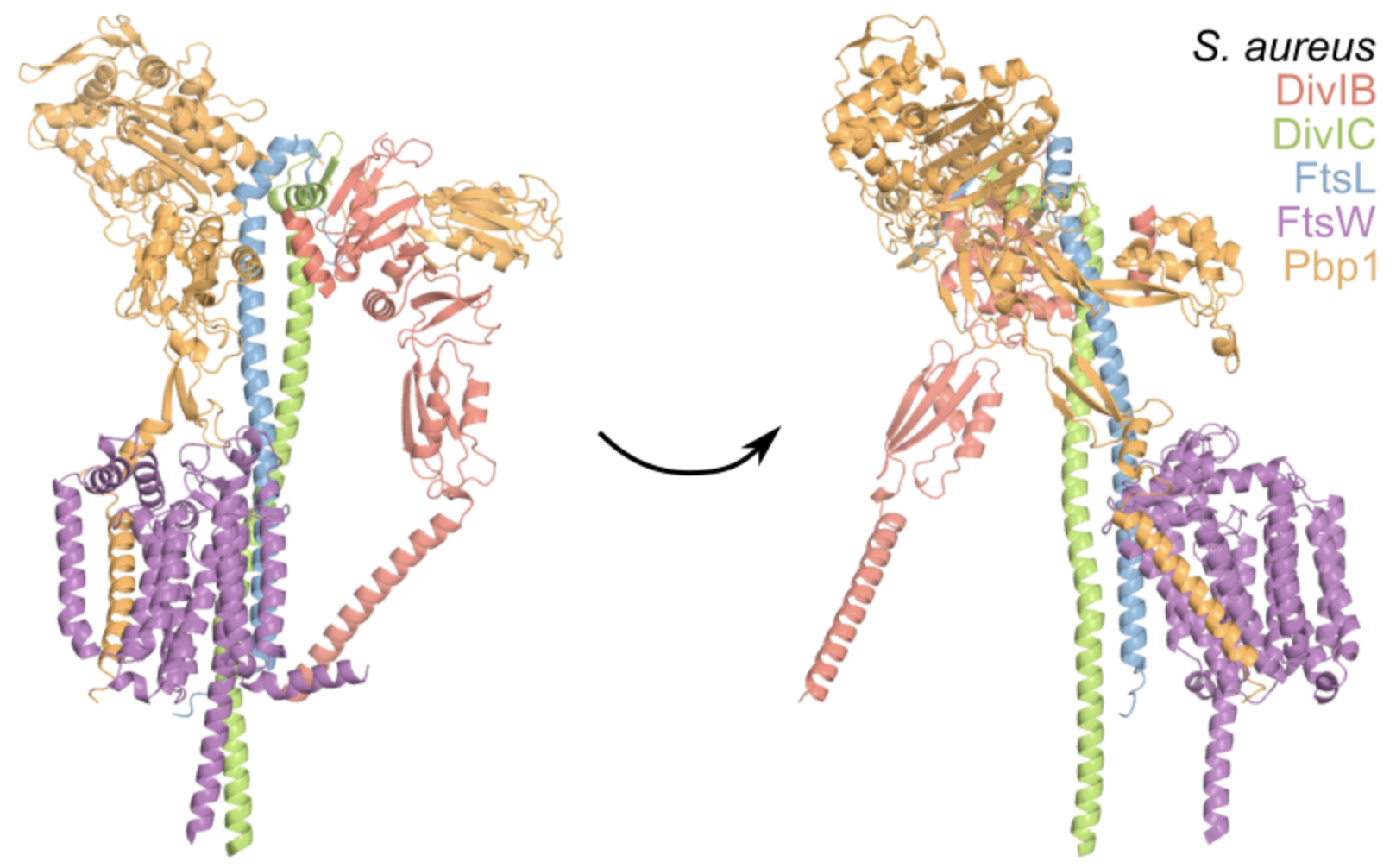
Abstract: Bacterial cell division requires recruitment of peptidoglycan (PG) synthases to the division site by the tubulin homologue, FtsZ. Septal PG synthases promote septum growth. FtsZ treadmilling is proposed to drive the processive movement of septal PG synthases and septal constriction in some bacteria; however, the precise mechanisms spatio-temporally regulating PG synthase movement and activity and FtsZ treadmilling are poorly understood. Here using single-molecule imaging of division proteins in the Gram-positive pathogen Staphylococcus aureus, we showed that the septal PG synthase complex FtsW/PBP1 and its putative activator protein, DivIB, move with similar velocity around the division site. Impairing FtsZ treadmilling did not affect FtsW or DivIB velocities or septum constriction rates. Contrarily, PG synthesis inhibition decelerated or stopped directional movement of FtsW and DivIB, and septum constriction. Our findings suggest that a single population of processively moving FtsW/PBP1 associated with DivIB drives cell constriction independently of FtsZ treadmilling in S. aureus.
S Schäper, AD Brito, BM Saraiva, GR Squyres, MJ Holmes, EC Garner, Z Hensel, R Henriques, MG Pinho*. Cell constriction requires processive septal peptidoglycan synthase movement independent of FtsZ treadmilling in Staphylococcus aureus. Nature Microbiology, 2024. https://doi.org/10.1038/s41564-024-01629-6

Abstract: Bacterial cell division requires recruitment of peptidoglycan (PG) synthases to the division site by the tubulin homologue, FtsZ. Septal PG synthases promote septum growth. FtsZ treadmilling is proposed to drive the processive movement of septal PG synthases and septal constriction in some bacteria; however, the precise mechanisms spatio-temporally regulating PG synthase movement and activity and FtsZ treadmilling are poorly understood. Here using single-molecule imaging of division proteins in the Gram-positive pathogen Staphylococcus aureus, we showed that the septal PG synthase complex FtsW/PBP1 and its putative activator protein, DivIB, move with similar velocity around the division site. Impairing FtsZ treadmilling did not affect FtsW or DivIB velocities or septum constriction rates. Contrarily, PG synthesis inhibition decelerated or stopped directional movement of FtsW and DivIB, and septum constriction. Our findings suggest that a single population of processively moving FtsW/PBP1 associated with DivIB drives cell constriction independently of FtsZ treadmilling in S. aureus.
# F Débarre*, Z Hensel. A critical reexamination of recovered SARS-CoV-2 sequencing data. bioRxiv 580500, 2024. DOI: https://doi.org/10.1101/2024.02.15.580500
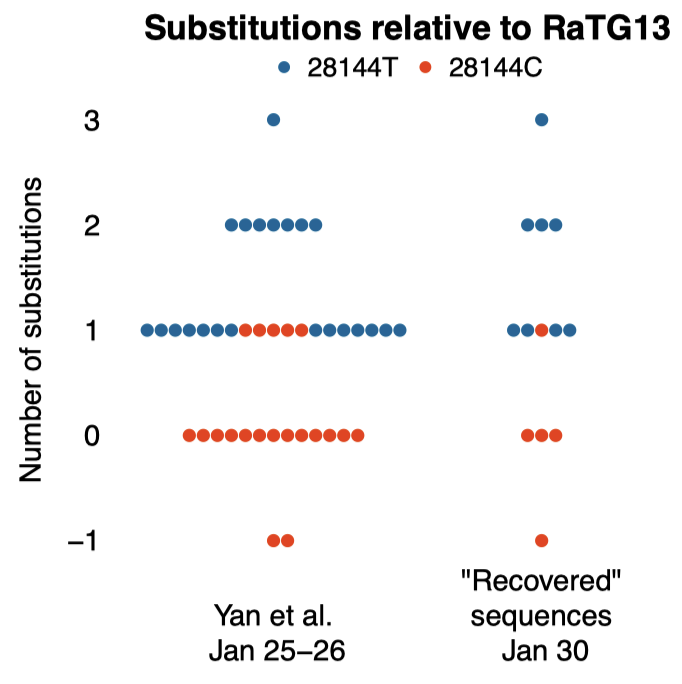
Abstract: SARS-CoV-2 genomes collected at the onset of the Covid-19 pandemic are valuable because they could help understand how the virus entered the human population. In 2021, Jesse Bloom reported on the recovery of a dataset of raw sequencing reads that had been removed from the NCBI SRA database at the request of the data generators, a scientific team at Wuhan University (Wang et al., 2020b). Bloom suggested that the data may have been removed in order to obfuscate the origin of SARS-CoV-2, and he questioned the generating authors’ statements that the samples had been collected on and after January 30, 2020. Here, we show that sample collection dates were published in 2020 by Wang et al. together with the sequencing reads, and match the dates given by the authors in 2021. We examine mutations in these sequences and confirm that they are entirely consistent with the previously known genetic diversity of SARS-CoV-2 of late January 2020. Finally, we explain how an apparent phylogenetic rooting paradox described by Bloom was resolved by subsequent analysis. Our reanalysis demonstrates that allegations of cover-up or of metadata manipulation were unwarranted.
L Zimmermann, X Zhao, J Makroczyova, M Wachsmuth-Melm, V Prasad, Z Hensel, R Bartenschlager, P Chlanda*. SARS-CoV-2 nsp3 and nsp4 are minimal constituents of a pore spanning replication organelle. Nature Communications 14, 2023. https://doi.org/10.1038/s41467-023-43666-5
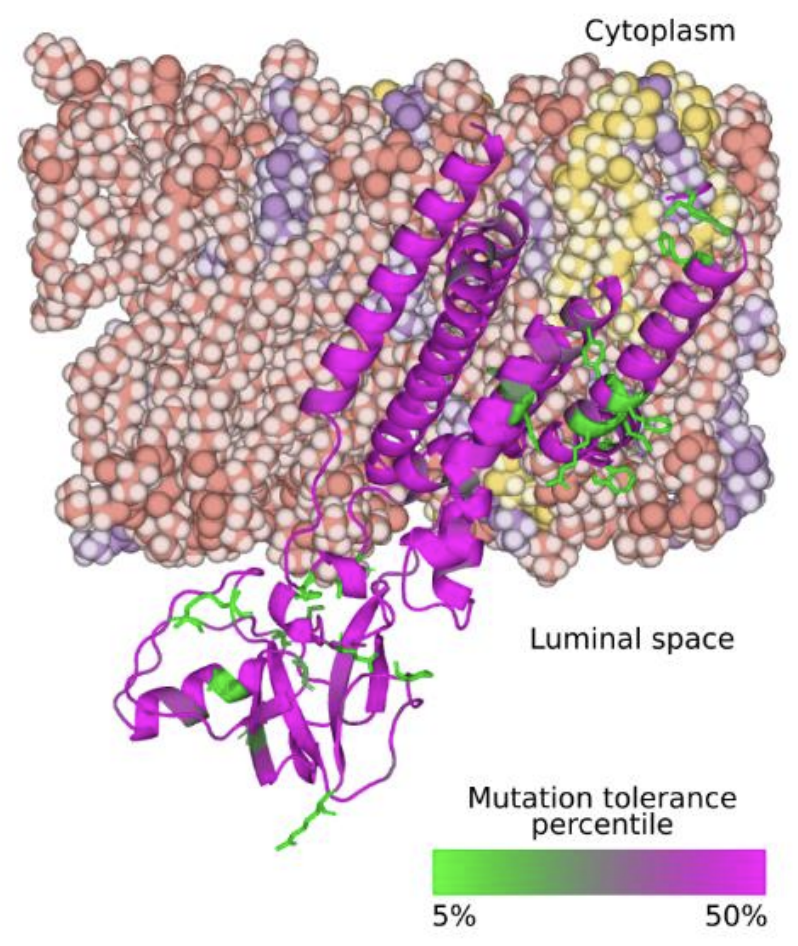
Abstract: Coronavirus replication is associated with the remodeling of cellular membranes, resulting in the formation of double-membrane vesicles (DMVs). A DMV-spanning pore was identified as a putative portal for viral RNA. However, the exact components and the structure of the SARS-CoV-2 DMV pore remain to be determined. Here, we investigate the structure of the DMV pore by in situ cryo-electron tomography combined with subtomogram averaging. We identify non-structural protein (nsp) 3 and 4 as minimal components required for the formation of a DMV-spanning pore, which is dependent on nsp3-4 proteolytic cleavage. In addition, we show that Mac2-Mac3-DPUP-Ubl2 domains are critical for nsp3 oligomerization and crown integrity which influences membrane curvature required for biogenesis of DMVs. Altogether, SARS-CoV-2 nsp3-4 have a dual role by driving the biogenesis of replication organelles and assembly of DMV-spanning pores which we propose here to term replicopores.
†BM Britton, †RA Yovanno, SF Costa, J McCausland, AY Lau, J Xiao*, Z Hensel*. Conformational changes in the essential E. coli septal cell wall synthesis complex suggest an activation mechanism. Nature Communications 14, 2023. https://doi.org/10.1038/s41467-023-39921-4
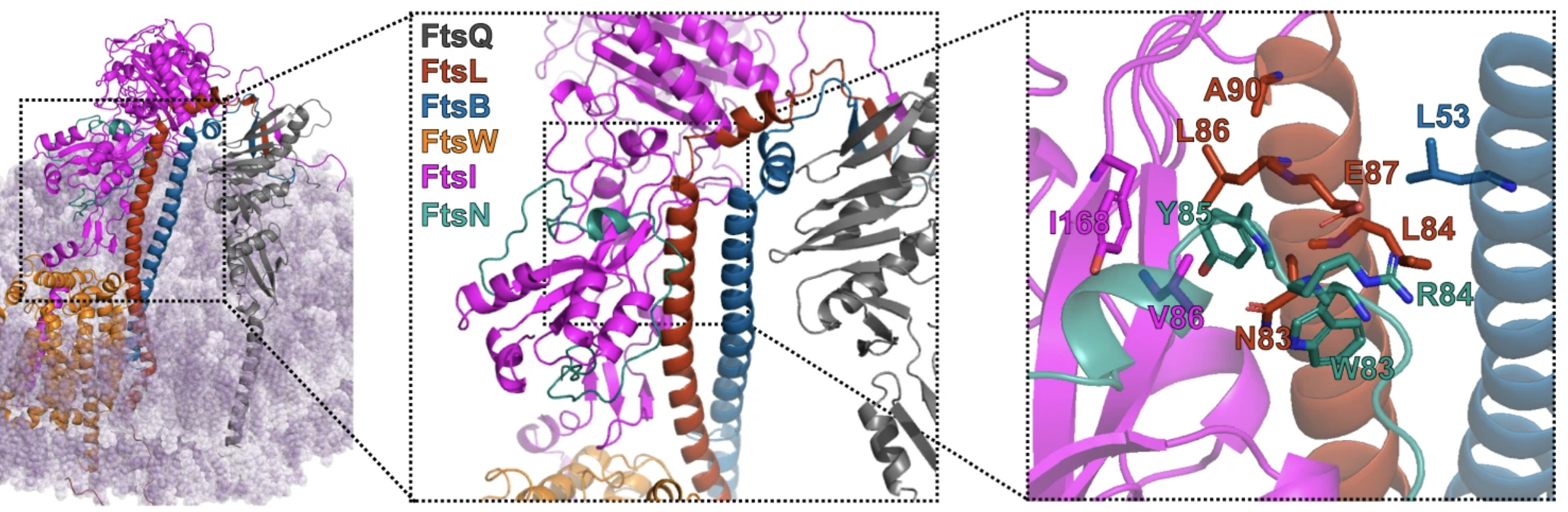
Abstract: The bacterial divisome is a macromolecular machine composed of more than 30 proteins that controls cell wall constriction during division. Here, we present a model of the structure and dynamics of the core complex of the E. coli divisome, supported by a combination of structure prediction, molecular dynamics simulation, single-molecule imaging, and mutagenesis. We focus on the septal cell wall synthase complex formed by FtsW and FtsI, and its regulators FtsQ, FtsL, FtsB, and FtsN. The results indicate extensive interactions in four regions in the periplasmic domains of the complex. FtsQ, FtsL, and FtsB support FtsI in an extended conformation, with the FtsI transpeptidase domain lifted away from the membrane through interactions among the C-terminal domains. FtsN binds between FtsI and FtsL in a region rich in residues with superfission (activating) and dominant negative (inhibitory) mutations. Mutagenesis experiments and simulations suggest that the essential domain of FtsN links FtsI and FtsL together, potentially modulating interactions between the anchor-loop of FtsI and the putative catalytic cavity of FtsW, thus suggesting a mechanism of how FtsN activates the cell wall synthesis activities of FtsW and FtsI.
# Z Hensel*. Predicted binding interface between coronavirus nsp3 and nsp4. bioRxiv 483145, 2022. https://doi.org/10.1101/2022.03.05.483145
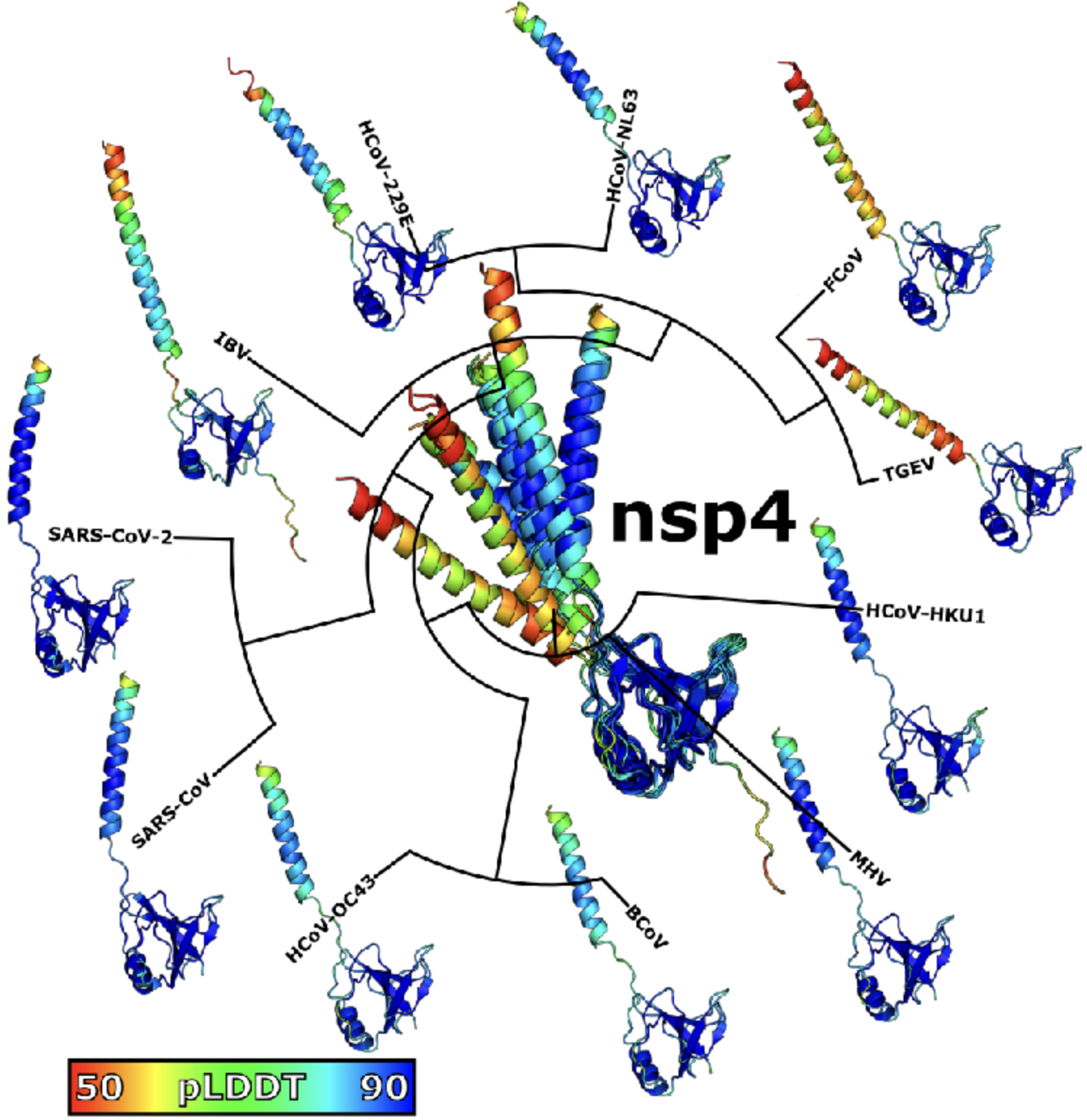
Abstract: Double membrane vesicles (DMVs) in coronavirus-infected cells feature pores that span both membranes. DMV pores were observed to have six-fold symmetry and include the nsp3 protein. Co-expression of SARS-CoV nsp3 and nsp4 induces DMV formation, and elements of nsp3 and nsp4 have been identified that are essential for membrane disruption. I describe a predicted luminal binding interface between nsp3 and nsp4 that is membrane-associated, conserved in SARS-CoV-2 during the COVID-19 pandemic and in diverse coronaviruses, and stable in molecular dynamics simulation. Combined with structure predictions for the full-length nsp4 monomer and cryo-EM data, this suggests a DMV pore model in which nsp4 spans both membranes with nsp3 and nsp4 inserted into the same bilayer. This approach may be able to identify additional protein-protein interactions between coronavirus proteins.
†R Letra-Vilela, †R Quiteres, F Murtinheira, A Crevenna, Z Hensel*, F Herrera*. New tools for the visualization of glial fibrillary acidic protein in living cells. Experimental Results 1, 2020. DOI: 10.1017/exp.2020.1.
JPN Silva, SV Lopes, DJ Grilo, Z Hensel*. Plasmids for independently tunable, low-noise expression of two genes. mSphere 4:e00340-19, 2019. DOI: 10.1128/mSphere.00340-19.
X Fang, Q Liu, C Bohrer, Z Hensel, W Han, J Wang, J Xiao*. New cell fate potentials and switching kinetics uncovered in a classic bistable genetic switch. Nature Communications 9, 2018. DOI: 10.1038/s41467-018-05071-1.
Z Hensel*. A plasmid-based Escherichia coli gene expression system with cell-to-cell variation below the extrinsic noise limit. PLoS ONE 12, 2017. DOI: 10.1371/journal.pone.0187259.
Z Hensel, TT Marquez-Lago. Cell-cycle-synchronized, oscillatory expression of a negatively autoregulated gene in E. coli. arXiv 1506.08596, 2015. Link.
Z Hensel, X Weng, AC Lagda, J Xiao*. Transcription-factor-mediated DNA looping probed by high-resolution, single-molecule imaging in live E. coli cells. PLOS Biology 11, 2013. DOI: 10.1371/journal.pbio.1001591.
†Z Hensel, †H Feng (equal contribution), B Han, C Hatem, J Wang, J Xiao*. Stochastic expression dynamics of a transcription factor revealed by single-molecule noise analysis. Nature Structural and Molecular Biology 19, 2012. DOI: 10.1038/nsmb.2336.
H Feng, Z Hensel, J Xiao, J Wang*. Analytical calculation of protein production distributions in models of clustered protein expression. Physical Review E 85, 2012. DOI: 10.1103/PhysRevE.85.031904.
G Fu, T Huang, J Buss, C Coltharp, Z Hensel, J Xiao*. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PLoS ONE 5, 2010. DOI: 10.1371/journal.pone.0012680.
E Barry, Z Hensel, Z Dogic, M Shribak, R Oldenbourg. Entropy-driven formation of a chiral liquid-crystalline phase of helical filaments. Physical Review Letters 96, 2006. DOI: 10.1103/PhysRevLett.96.018305.
Reviews/Highlights
Z Hensel, J Xiao*. Single-molecule methods for studying gene regulation in vivo. Pflügers Archiv-European Journal of Physiology 465, 2013. DOI: 10.1007/s00424-013-1243-y.
Z Hensel, J Xiao*. A mechanism for stochastic decision making by bacteria. ChemBioChem 10, 2009. DOI: 10.1002/cbic.200800824.
Correspondence
S Sarabipour*, HJ Debat, E Emmott, SJ Burgess, B Schwessinger, Z Hensel. On the value of preprints: an early career researcher perspective. PLoS Biology 17(2): e3000151, 2019. DOI: 10.1371/journal.pbio.3000151.
S Sarabipour*, EM Wissink, SJ Burgess, Z Hensel, HJ Debat, E Emmott, A Akay, KC Akdemir, B Schwessinger. Preprints are good for science and good for the public. Nature 560, 2018. DOI: 10.1038/d41586-018-06054-4.
†S Sarabipour*, †EM Wissink*, †SJ Burgess*, Z Hensel, H Debat, E Emmott, A Akay, KC Akdemir, B Schwessinger. Maintaining confidence in the reporting of scientific outputs. PeerJ Preprints 6, 2018. DOI: 10.7287/peerj.preprints.27098v1.
Patents/Methods
J Xiao, Z Hensel. Co-translational activation of a transcription factor by proteolytic cleavage and methods of use. US Patent 9,822,392 2017. Link.
Z Hensel, X Fang, J Xiao. Single-molecule imaging of gene regulation in vivo using cotranslational activation by cleavage (CoTrAC). Journal of Visualized Experiments, 2013. DOI: 10.3791/50042.
Published preprints
†JR Ferreira, †R Xu, Z Hensel*. Mycobacterium tuberculosis FtsB and PerM interact via a C-terminal helix in FtsB to modulate cell division. bioRxiv 584518, 2024. DOI: 10.1101/2024.03.11.584518.
Z Hensel*. Secondary structure of the SARS-CoV-2 genome is predictive of nucleotide substitution frequency. bioRxiv 581995, 2024. DOI: 10.1101/2024.02.27.581995.
A Crits-Christoph, JI Levy, JE Pekar, SA Goldstein, R Singh, Z Hensel, K Gangavarapu, MB Rogers, N Moshiri, RF Garry, EC Holmes, MPG Koopmans, P Lemey, S Popescu, A Rambaut, DL Robertson, MA Suchard, JO Wertheim, AL Rasmussen, KG Andersen, M Worobey*, F Débarre*. *Genetic tracing of market wildlife and viruses at the epicenter of the COVID-19 pandemic. bioRxiv 557637, 2023. DOI: 10.1101/2023.09.13.557637
S Schäper, AD Brito, BM Saraiva, GR Squyres, MJ Holmes, EC Garner, Z Hensel, R Henriques, MG Pinho*. Processive movement of Staphylococcus aureus essential septal peptidoglycan synthases is independent of FtsZ treadmilling and drives cell constriction. bioRxiv 547026, 2023. DOI:10.1101/2023.06.29.547026
†BM Britton, †RA Yovanno, SF Costa, J McCausland, AY Lau, J Xiao*, Z Hensel*. Conformational changes in the essential E. coli septal cell wall synthesis complex suggest an activation mechanism. bioRxiv 518129, 2022 DOI: 10.1101/2022.11.27.518129
JPN Silva, SV Lopes, DJ Grilo, Z Hensel*. Plasmids for independently tunable, low-noise expression of two genes. bioRxiv 515940, 2019. DOI: 10.1101/515940.
S Sarabipour*, HJ Debat, E Emmott, SJ Burgess, B Schwessinger, Z Hensel. On the value of preprints: an early career researcher perspective. PeerJ Preprints 6, 2018. DOI: 10.7287/peerj.preprints.27400v1.
X Fang, Q Liu, C Bohrer, Z Hensel, W Han, J Wang, J Xiao*. New cell fate potentials and switching kinetics uncovered in a classic bistable genetic switch. bioRxiv 215061, 2017. DOI: 10.1101/215061.
Z Hensel*. A plasmid-based Escherichia coli gene expression system with cell-to-cell variation below the extrinsic noise limit. bioRxiv 192963, 2017. DOI: 10.1101/192963.
E Barry, Z Hensel, M Shribak, R Oldenbourg, Z Dogic. Entropy-driven formation of a chiral liquid-crystalline phase of helical filaments. arXiv cond-mat/0510708, 2005. Link.
