Radio interview/podcast about new preprint
April 24, 2025
Zach was interviewed with co-author Florence Débarre (Sorbonne) about a new preprint we published on early SARS-CoV-2 sequence diversity for BBC News’ Science in Action. Check out the beginning of the podcasts for our comments on the preprint and stick around until the end to learn about a gruesome carnivorous caterpillar.
Here is a summary of the beginning of the program:
Wet market SARS CoV-2 origins revisited
Last week, the website covid.gov looked very different, containing information on coping with covid and US research. This week it leads you to a White House webpage outlining the lab-leak hypothesis – that the pandemic was the result of dodgy lab work at the Wuhan Institute of Virology. The scientific consensus however continues to suggest a zoonotic crossover event. And a preprint recently published by Zach Hensel of ITQB NOVA (NOVA University Lisbon) and Florence Débarre of the Sorbonne, adds new early sequences to the evidence base. As they suggest, it doesn’t seal the debate, but it certainly doesn’t surprise the scientists.
view this post on its own page
Paper out on mycobacterial cell division
March 26, 2025
Work from our lab by João Ramalheira Ferreira, Ruilan Xu, and Zach Hensel is available online today in Journal of Bacteriology!
Our research reveals significant insights into the dynamic interaction between the proteins PerM and FtsB within Mycobacterium tuberculosis, contributing to our understanding of bacterial cell division mechanisms crucial for infection persistence. By combining innovative fluorescence microscopy and molecular dynamics, we established that the stability of these proteins is interdependent; molecular dynamics placing PerM-FtsB in the context of the mycobacterial divisome shows how disrupting PerM-FtsB interactions can plausibly impact bacterial cell wall synthesis. These findings highlight the PerM-FtsB interface as a promising target for novel therapeutics aimed at combating persistent bacterial infections. Importantly, our approach can be adapted for similar studies in other bacterial systems, suggesting broad implications for microbial biology and antibiotic development.
view this post on its own page
New paper published on pandemic origins
September 17, 2024
A paper co-authored with Crits-Cristoph and colleagues on environmental sampling at the epicenter of the COVID-19 pandemic has been published: Genetic tracing of market wildlife and viruses at the epicenter of the COVID-19 pandemic. We describe how metatranscriptomics data can be used to identify animals and animal viruses in Huanan Seafood Wholesale Market. Zach helped analyze statistical correlations in the data and commented on the manuscript for New Scientist:
Team member Zach Hensel at NOVA University Lisbon, Portugal, says the study highlights the preventable risks posed by the human-wildlife interface in Huanan market, and the need to mitigate these risks in similar locations.
“Even though humans were, of course, everywhere in Huanan market and live mammals were concentrated in a small number of stalls, human viruses, other than SARS2, were rare and several animal viruses were quite abundant,” says Hensel.
“This included one influenza virus with zoonotic spillover potential and an animal virus closely related to others that had caused devastating animal outbreaks.”
view this post on its own page
Posters presented at SMLMS
August 29, 2024
Ruilan and Zach presented posters at the Single-Molecule Localization Microscopy Symposium in Lisbon. Ruilan presented her work building on our recently updated preprint on studying an M. tuberculosis protein-protein interaction in the context of E. coli cells in order to observe the two proteins without other Mtb cell division proteins. Zach presented a summary of our progress towards efficiently counting single mRNAs as they are transcribed and quantifying transcription elongation rates for single mRNAs. This included Margarida Perreira’s work in her undergraduate research project in the lab testing whether newly reported fluorescent proteins mStayGold and mBaoJin improved upon mNeonGreen for mRNA imaging.
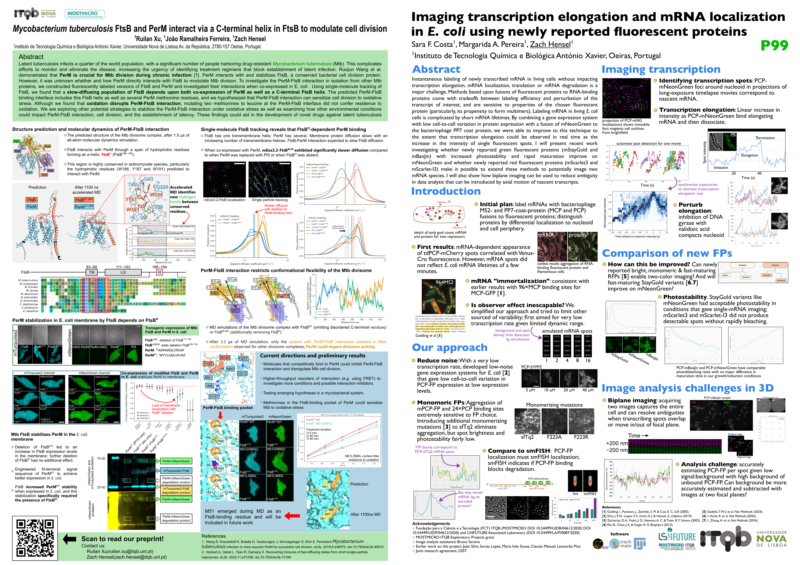
view this post on its own page
Recent work published from our lab
March 13, 2024
We’ve updated the research page to include updates on new preprints from our lab and in collaboration with other labs. Congrats to João and Ruilan for their work on the new preprint: Mycobacterium tuberculosis FtsB and PerM interact via a C-terminal helix in FtsB to modulate cell division. Congrats to lab alumnus Sara Costa as well for her paper from Mariana Pinho’s lab published last month: The role of GpsB in Staphylococcus aureus cell morphogenesis
We’ve also got two preprints out on SARS-CoV-2 related topics. First, with Florence Débarre, we found that stories about deleted viral sequences from June 2021 were not at all consistent with data available at the time. Read about it in the preprint: A critical reexamination of recovered SARS-CoV-2 sequencing data. We also published a very short preprint regarding the surprising observation that mutation rates in SARS-CoV-2 during the pandemic are strongly correlated with RNA basepairing. It will be interesting to see how this can be incorporated into models predicting the impact of mutations. Read the preprint here: Secondary structure of the SARS-CoV-2 genome is predictive of nucleotide substitution frequency.
view this post on its own page

