home — research — team — resources — lab news
Bem vindo!
Welcome to our lab! We are part of the MostMicro research unit at ITQB NOVA, an institute of Universidade NOVA de Lisboa. The lab started in January 2017.
Please contact zach.hensel@itqb.unl.pt if you are interested in working on a project in the lab or collaborating. Two of the possible themes for a research project are imaging transcription elongation in real time with single-molecule resolution and investigating how mycobacterial cell division responds to environmental stress both with fluorescence microscopy and computational methods.
Mission Statement
Our lab answers questions in microbial molecular biology, with a focus on single-molecule fluorescence microscopy in living cells. By detecting, tracking and counting individual DNA, mRNA and protein molecules, we will learn how to make better synthetic genetic circuits, how gene expression is regulated, and how cells interact with each other, other organisms, and their environments. We are especially interested in how single-cells evolve over time: our experiments complement modern high-resolution and high-throughput approaches that look at single time points. We closely follow developments in microscopy (especially emerging 3D imaging techniques) and fluorescent labeling. The lab embraces the open science revolution, with strong preferences for open access publication, data/code availability, and preprint.
Current research
Latest peer reviewed paper
From João Ramalheira Ferreira and Ruilan Xu in bioRxiv: Mycobacterium tuberculosis FtsB and PerM interact via a C-terminal helix in FtsB to modulate cell division
We used fluorescence microscopy including single-molecule tracking to investigate the M. tuberculosis PerM-FtsB interaction, expressing the proteins in E. colicells.
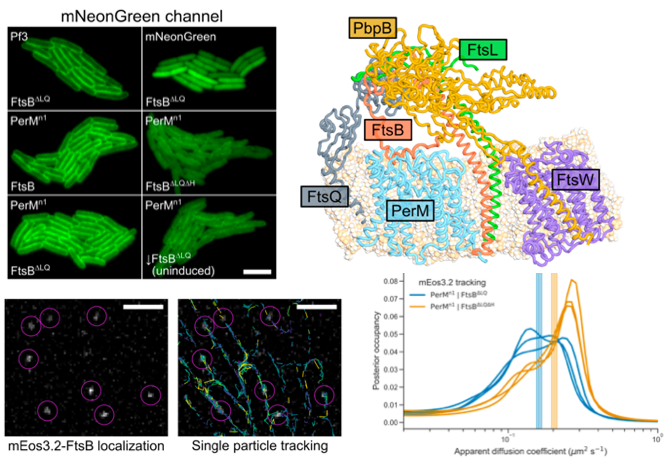
Here’s a movie of the M. tuberculosis divisome with PerM-FtsB interaction:
Other research in the lab
Check out our other recent work on a protein complex essential for remodelling the E. coli cell wall every cell time the cell divides. In collaboration with Jie Xiao's lab and Albert Lau's lab, we investigated the structure and dynamics of this “divisome” to examine how variations at key protein-protein interfaces could impact essential enzymatic activities.
Also see our low-noise gene expression system in E. coli (plasmids available at AddGene). We developed this to facilitate single-molecule experiments with fluorescent proteins that require very low expression levels with very low cell-to-cell variation. In João Silva’s thesis work, we subsequently extended this for independent tuning of low-noise expression of two proteins from two plasmids.
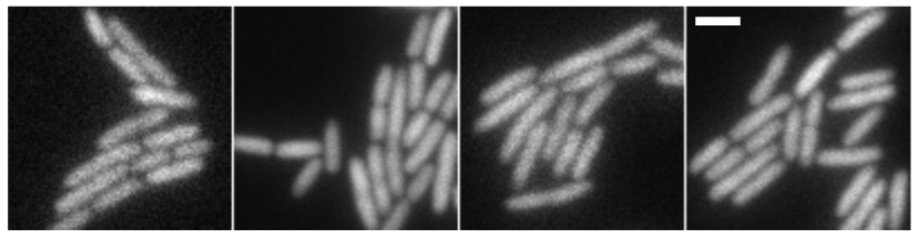 Gene expression from 100 to 10,000 molecules/cell with low cell-to-cell variation
Gene expression from 100 to 10,000 molecules/cell with low cell-to-cell variation
We are using this system to develop and improve methods for live-cell mRNA imaging. Browse the research page for details about our other projects.
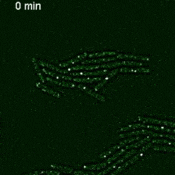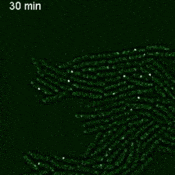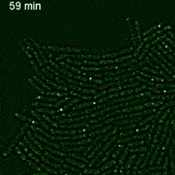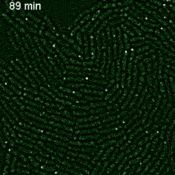 Imaging single mRNAs in E. coli
Imaging single mRNAs in E. coli
Funding
Our work is financially supported by the Fundação para a Ciência e a Tecnologia (FCT) through MOSTMICRO-ITQB (UIDB/04612/2020, UIDP/04612/2020) and LS4FUTURE (LA/P/0087/2020).
